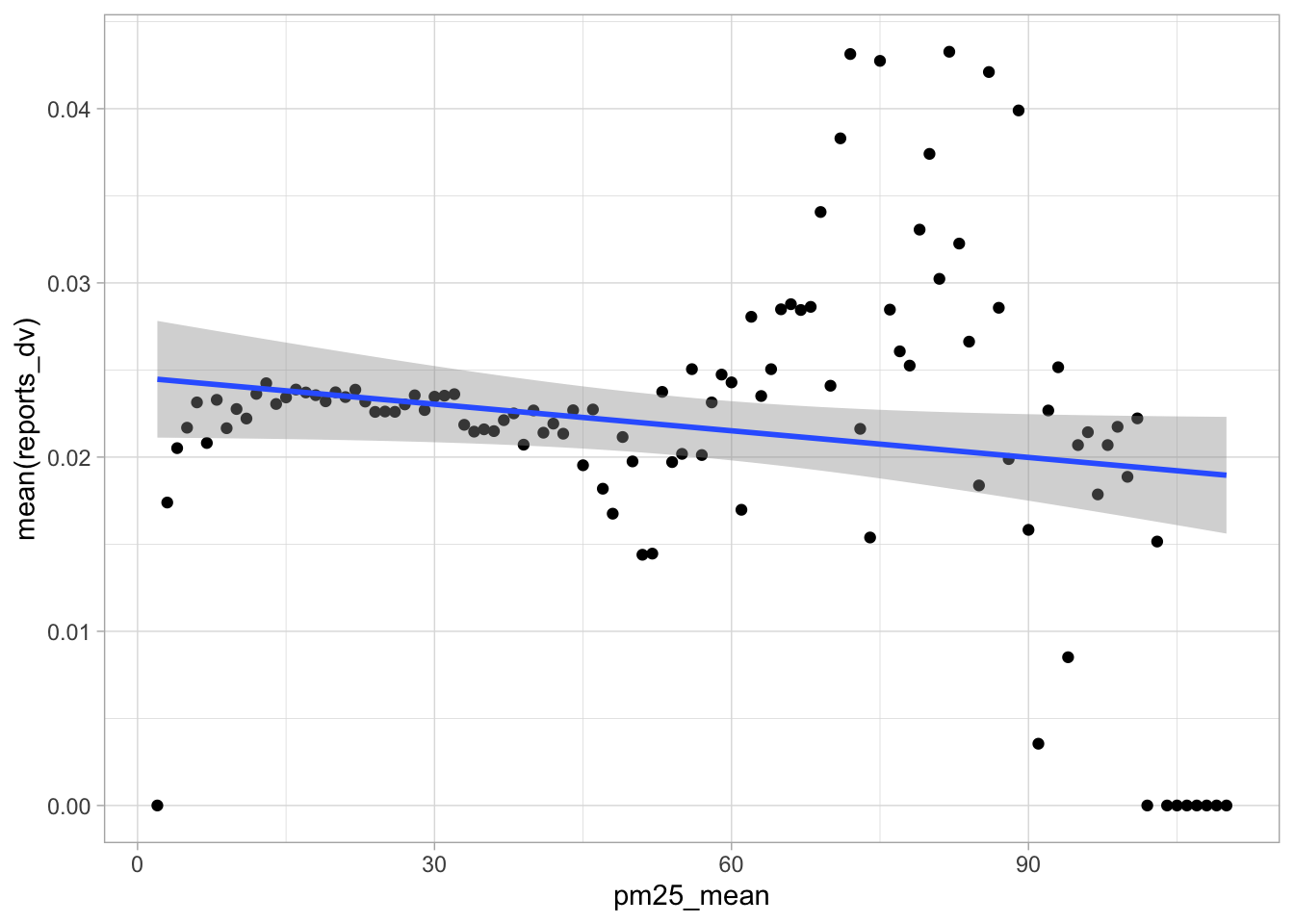
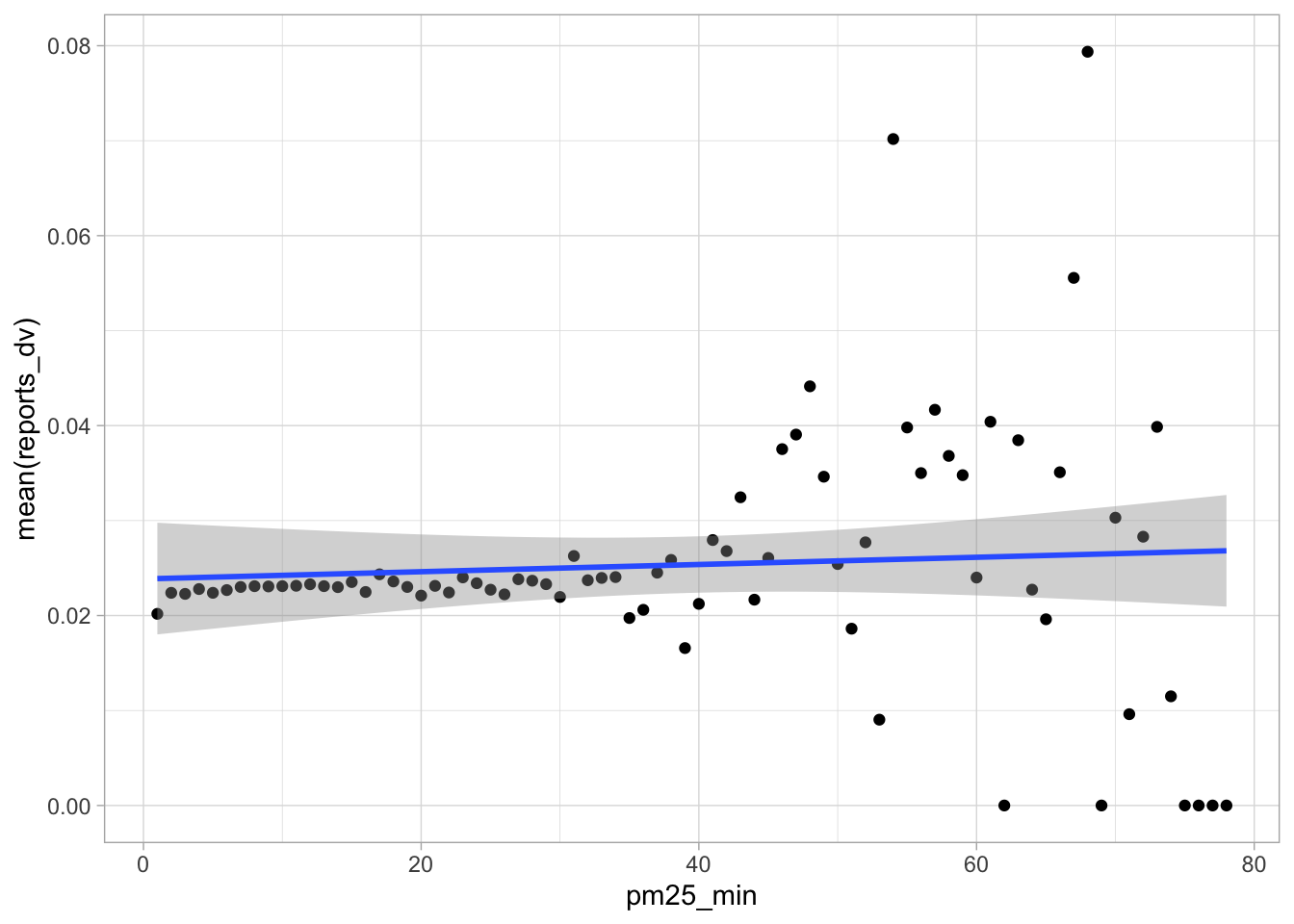
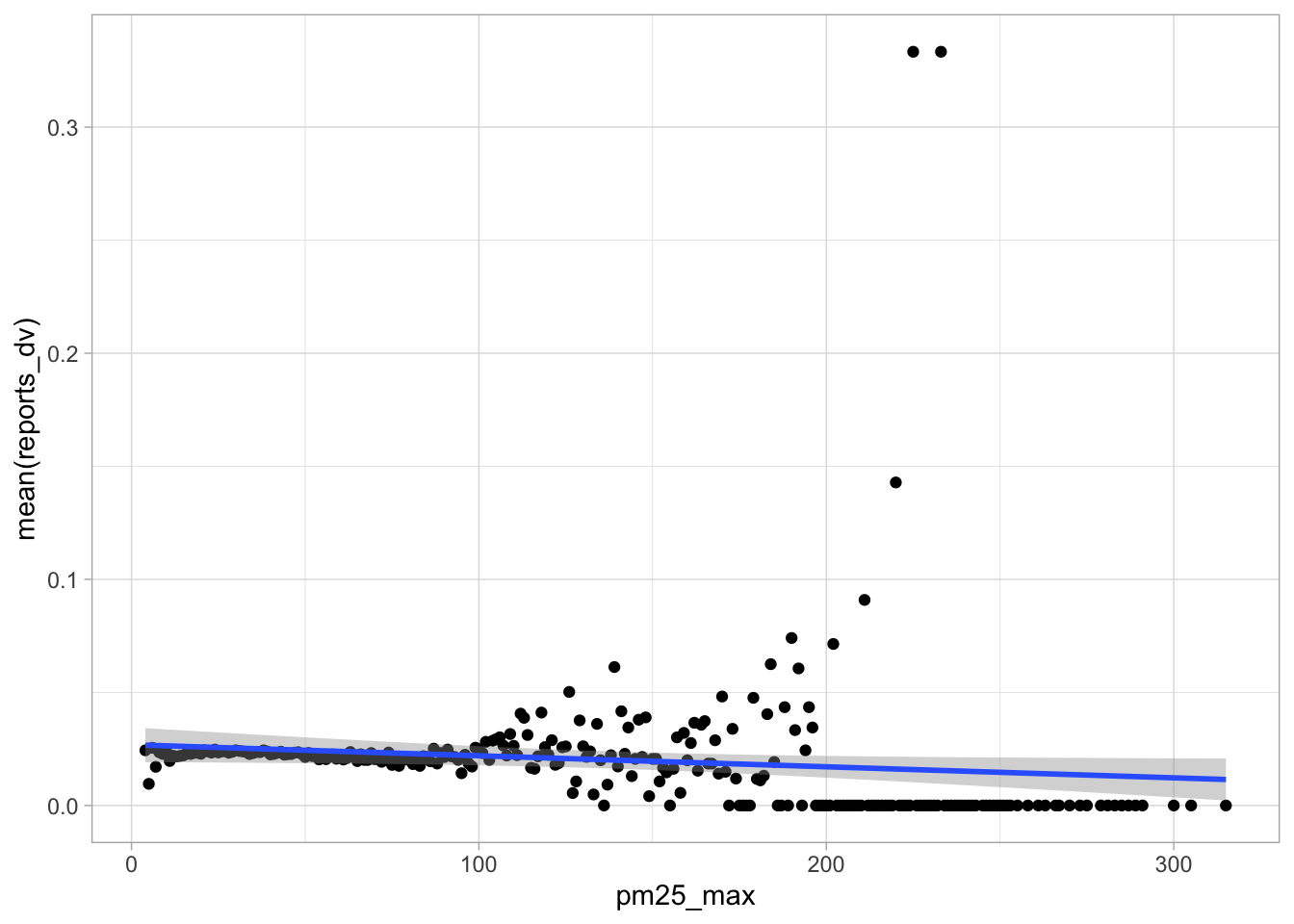
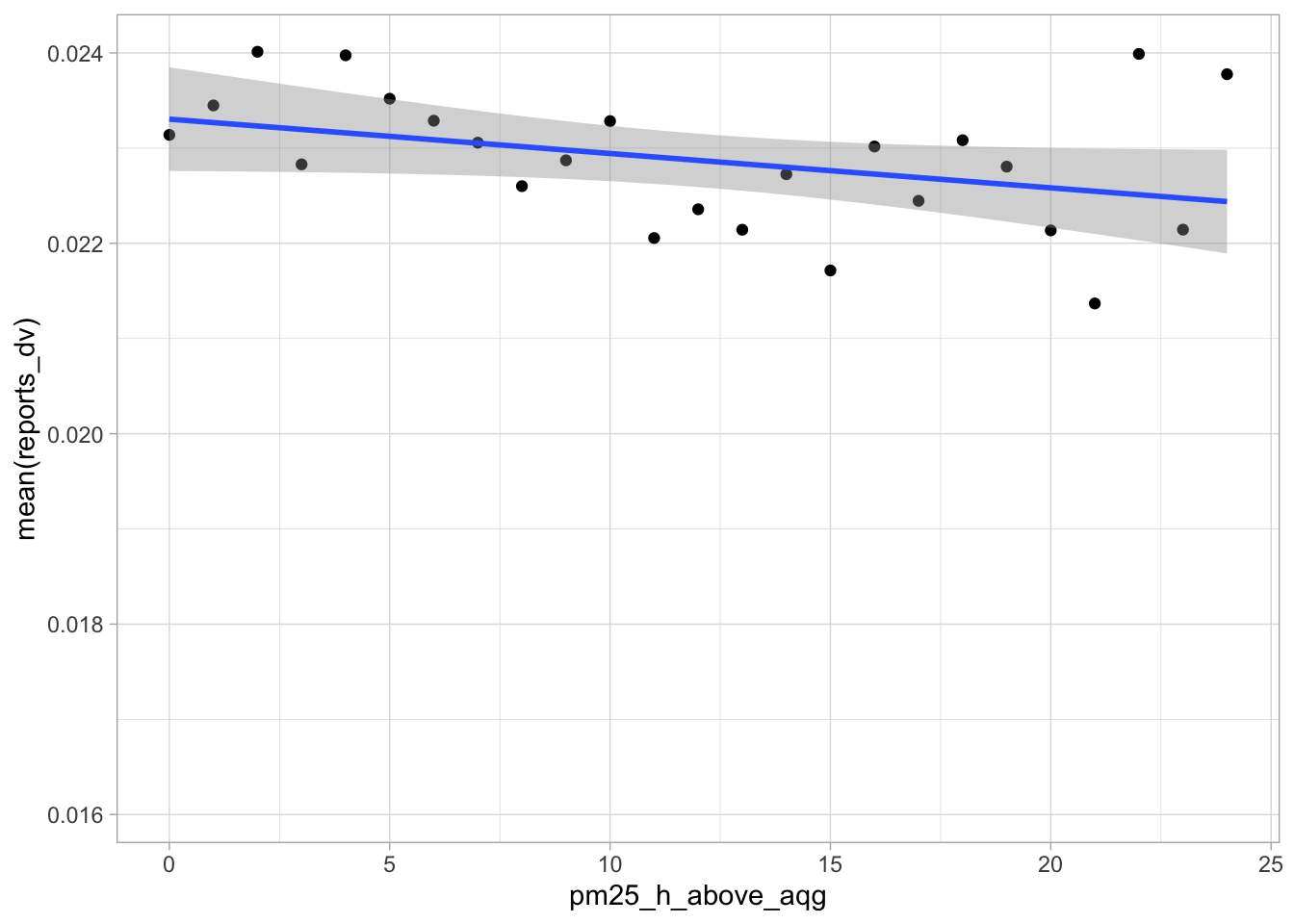
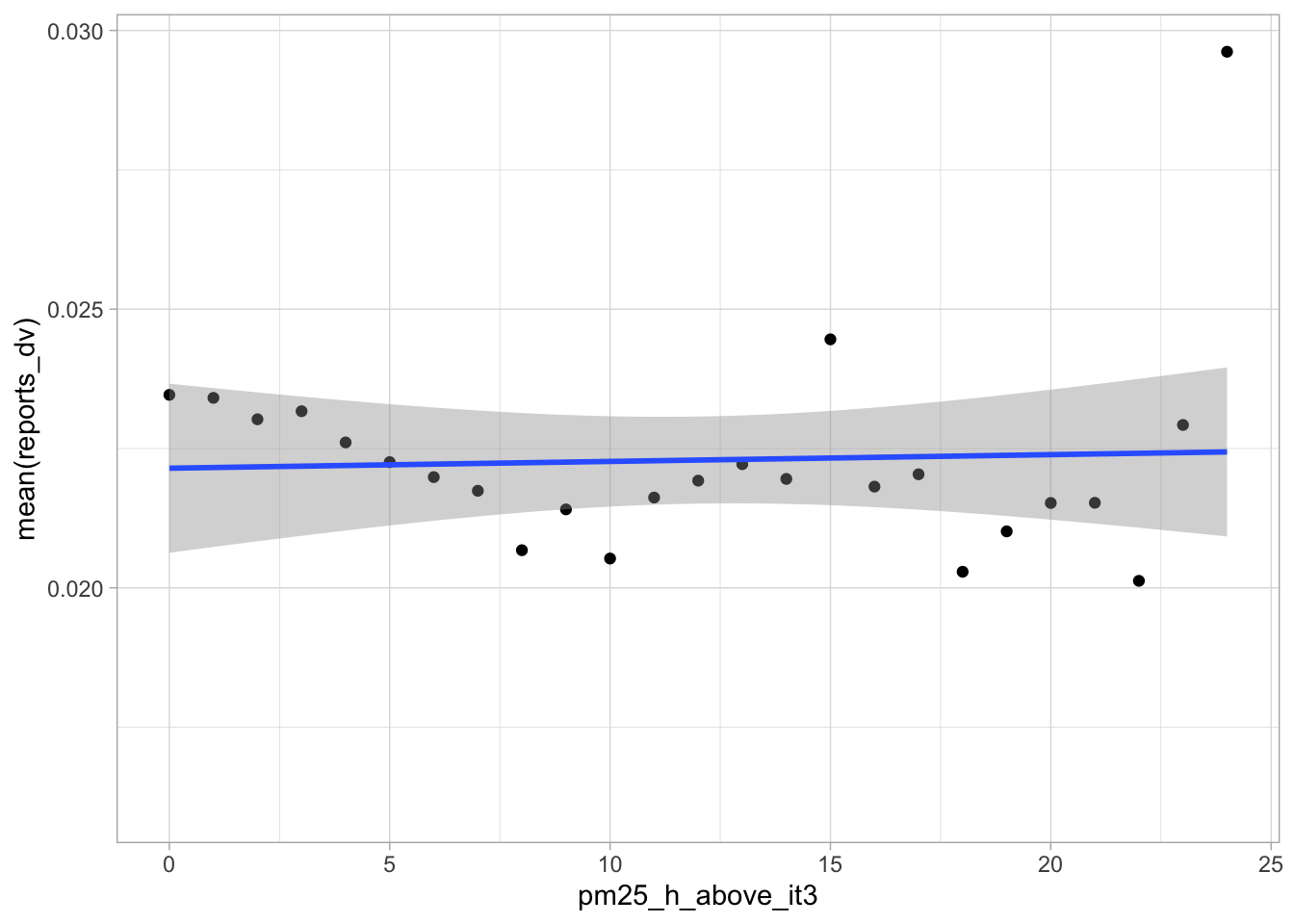
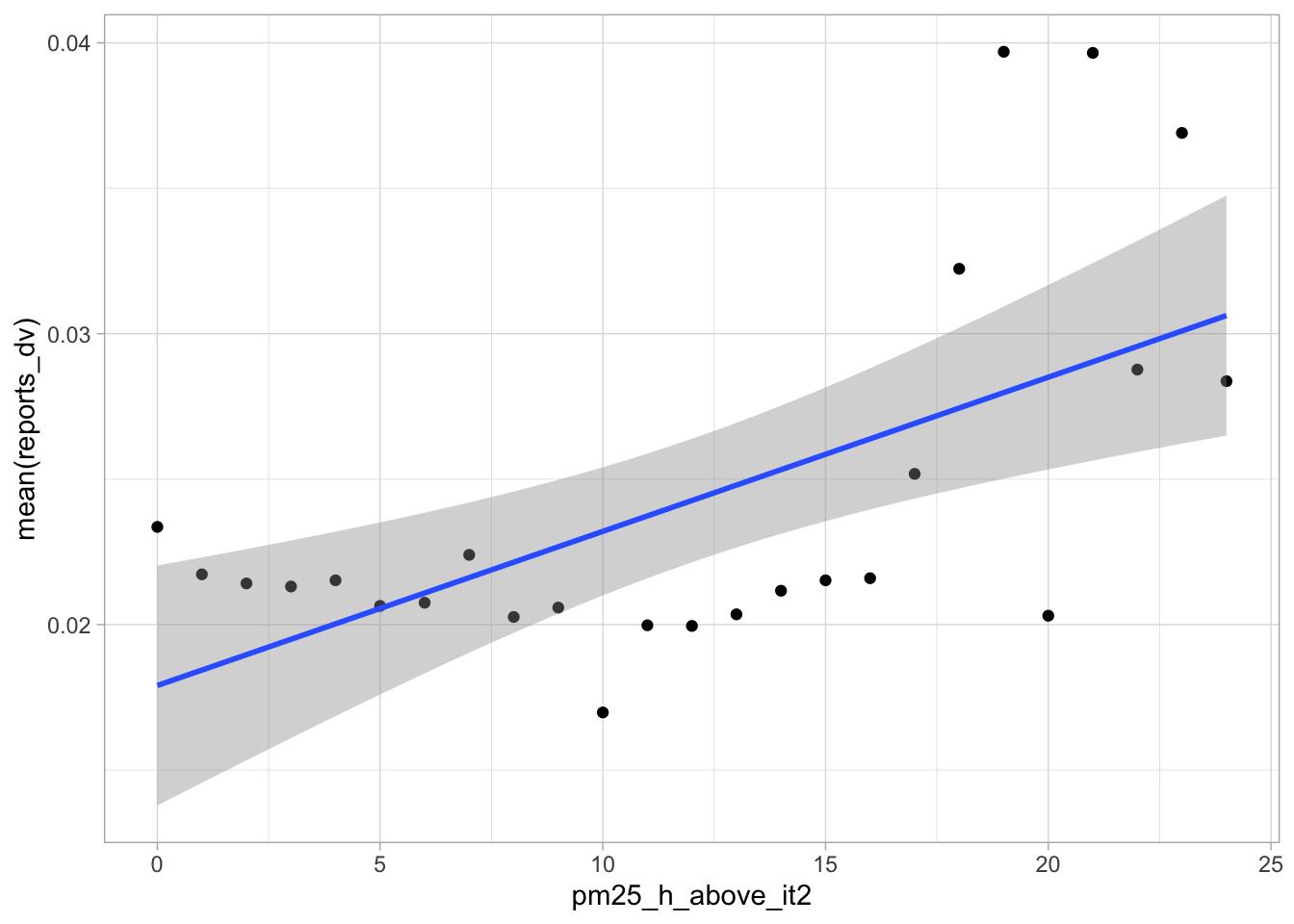
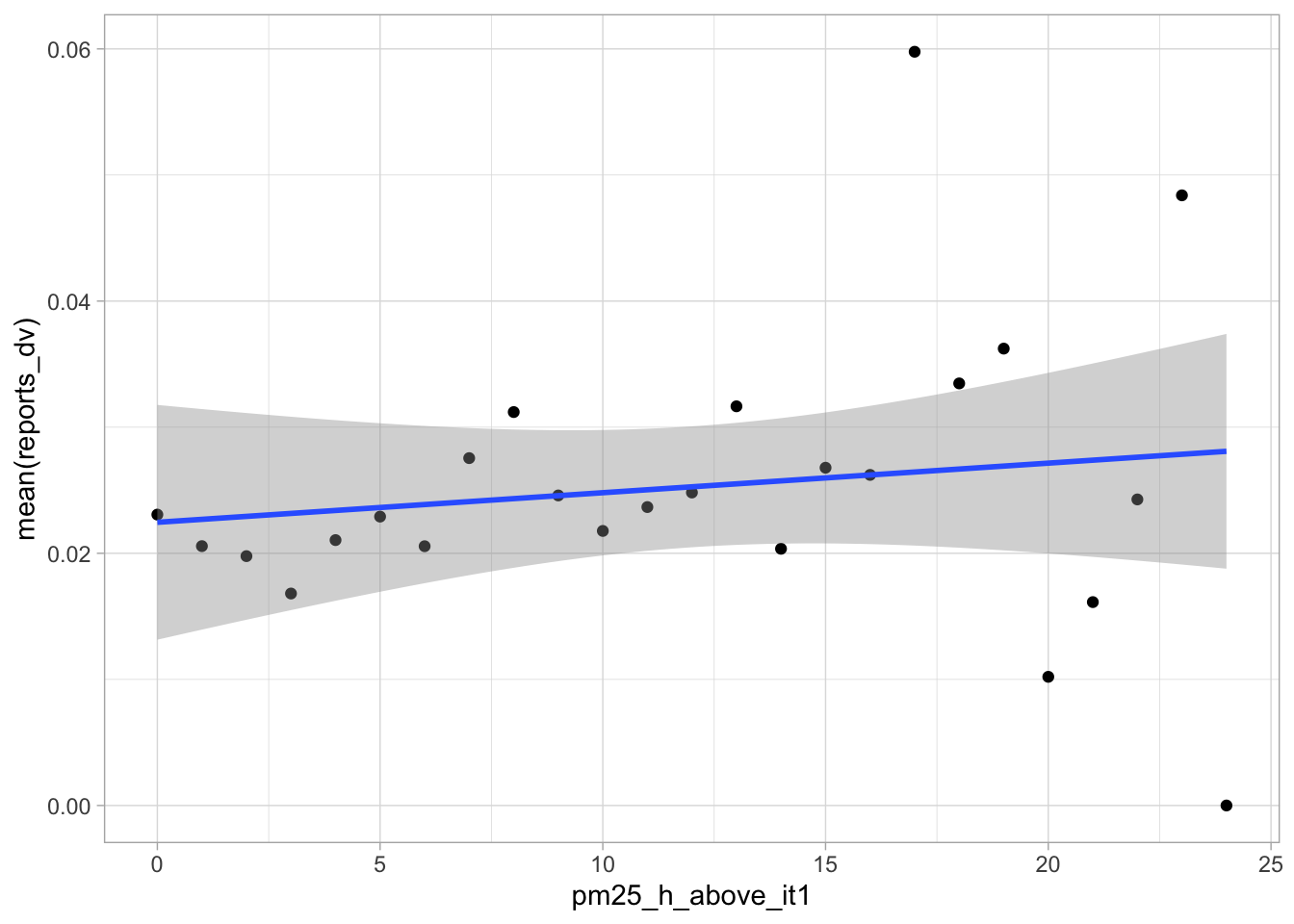
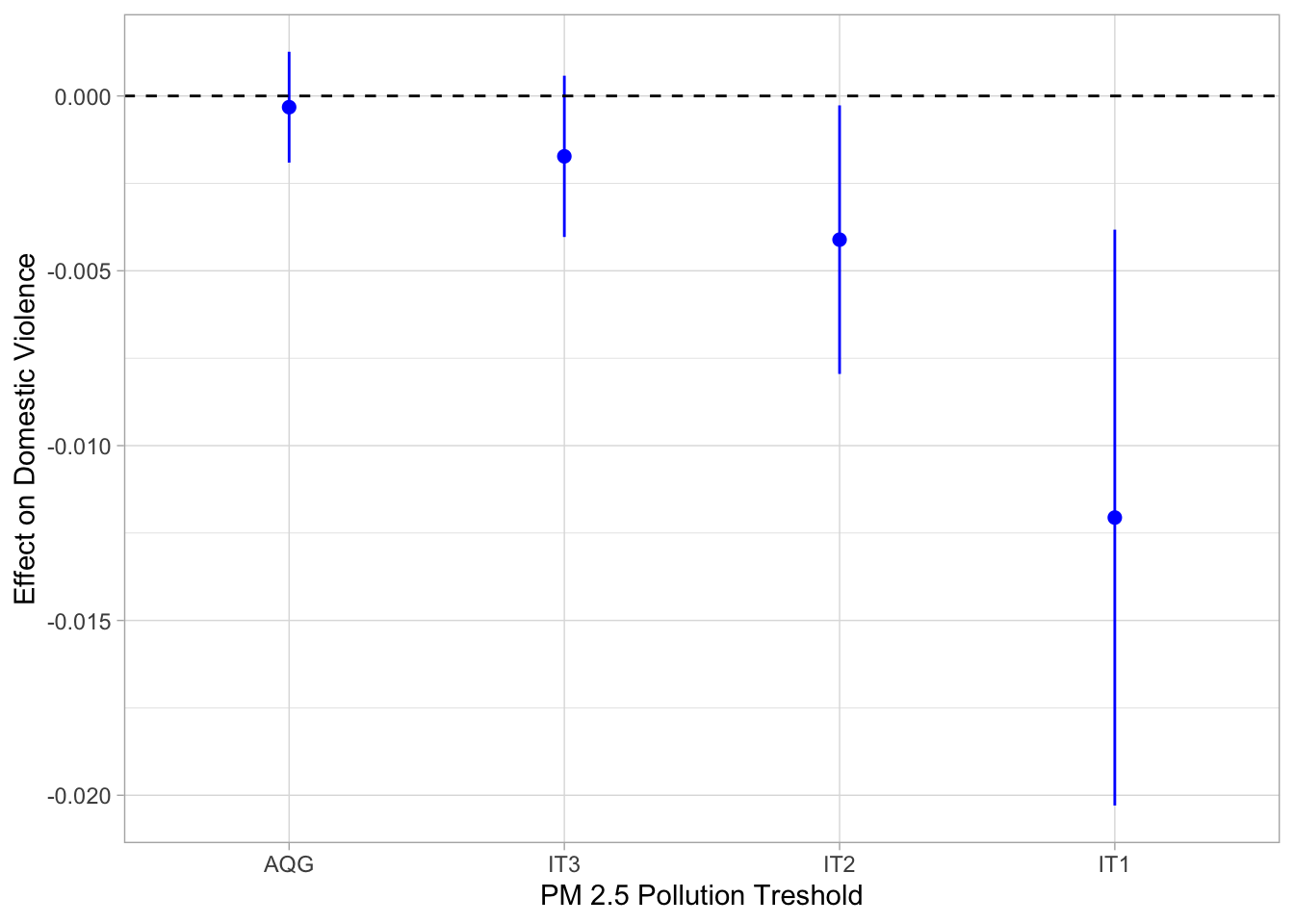
start_time <- Sys.time()
library(tidyverse)
library(here) # path control
library(lubridate) # handle dates
library(fixest) # estimations with fixed effects
library(modelsummary) # for the modelplot figure
library(tictoc)
library(knitr)
library(patchwork)
tic("Total render time")
mem.maxVSize(vsize = 40000)[1] 40000