start_time <- Sys.time()
library(tidyverse)
library(here) # path control
library(fixest) # estimations with fixed effects
library(knitr)
library(patchwork)
library(tictoc)
tic("Total render time")Main linear regressions
In this file I run the main regressions with the different measures of temperature and reports data.
- I export “Table 1”
- I save the plots with the estimates for the quintiles of precipitation, humidity and windspeed.
data <-
read_rds(here("..", "output", "data_reports.rds")) %>%
filter(covid_phase == "Pre-pandemic")
# quintiles are computed last minute, on the selected period
data <- data %>%
mutate(prec_quintile = ntile(prec, 5),
rh_quintile = ntile(rh, 5),
wsp_quintile = ntile(wsp, 5))
data <- data %>%
left_join(
read_rds(here("..", "output", "temporary_data",
"tmp_hourly_ageb.rds"))
)Run the regressions
Tmean
First, run the baseline regression with tmean, 24-hour mean of temperature
reg_linear_reports <- data %>%
fepois(reports_dv ~ tmean |
prec_quintile + rh_quintile + wsp_quintile +
ageb + year^month + day_of_week + day_of_year,
cluster = ~ ageb)Tmin and Tmax
Use tmin and tmax instead
reg_min <- data %>%
fepois(reports_dv ~ tmin |
prec_quintile + rh_quintile + wsp_quintile +
ageb + year^month + day_of_week + day_of_year,
cluster = ~ ageb)
reg_max <- data %>%
fepois(reports_dv ~ tmax |
prec_quintile + rh_quintile + wsp_quintile +
ageb + year^month + day_of_week + day_of_year,
cluster = ~ ageb)Or both in the same reg:
reg_min_max <- data %>%
fepois(reports_dv ~ tmin + tmax |
prec_quintile + rh_quintile + wsp_quintile +
ageb + year^month + day_of_week + day_of_year,
cluster = ~ ageb)Day and night temperatures
Max temperature is usually reached at 4-5pm
# max hour is always during the daytime
data %>%
filter(!is.na(tmax)) %>%
select(ageb, date, tmp_hourly) %>%
# indicator for which hour is the min in a day
mutate(max_hour = map_dbl(tmp_hourly, ~ which.max(.x))) %>%
ggplot() +
geom_histogram(aes(x = max_hour), bins = 24)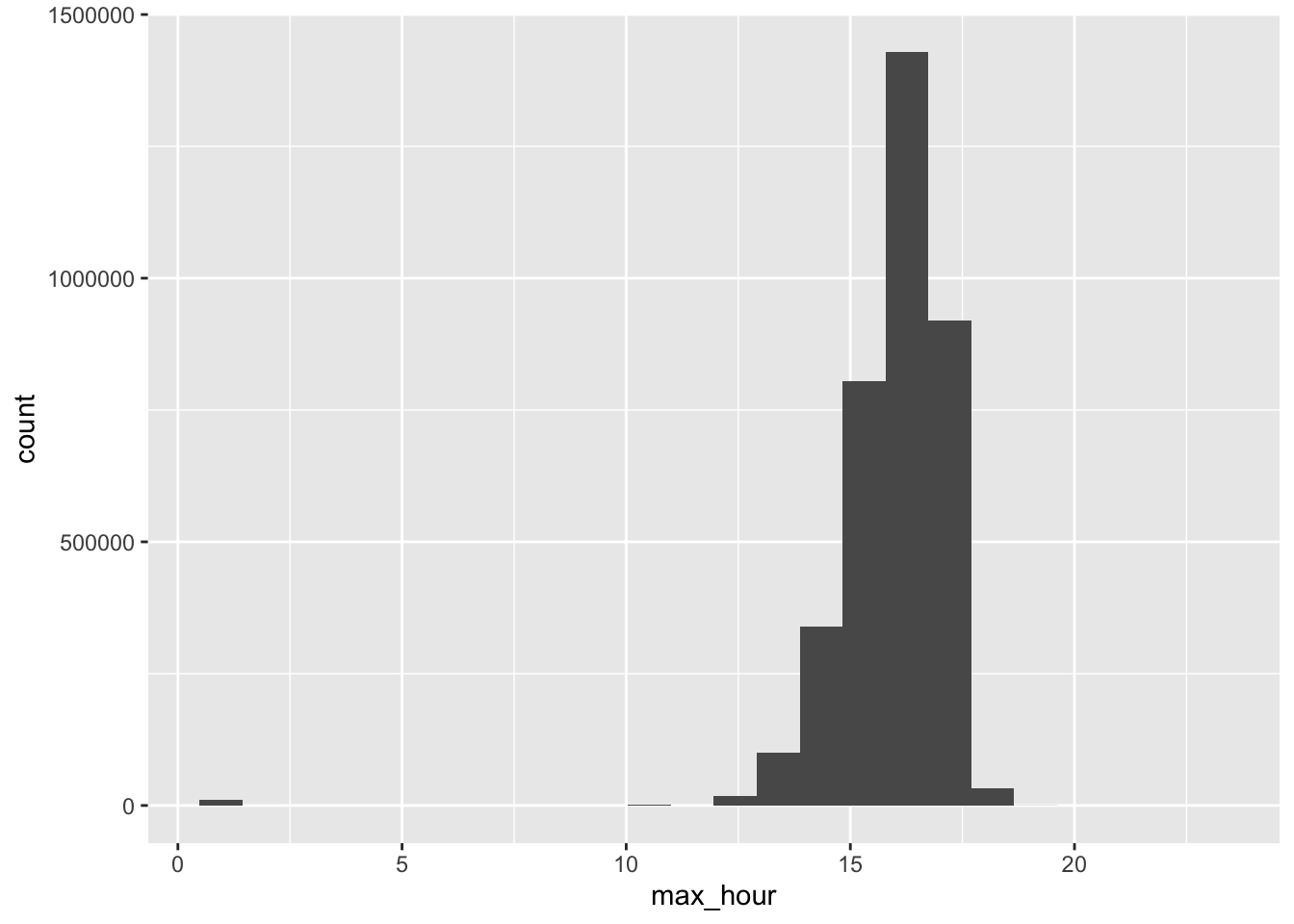
Min temperature is usually reached at hour 7, i.e from 06:00 to 06:59.
# hour in which the temp reaches its min is sometimes during daytime (hour 8, from 07:00 to 07:59)
data %>%
filter(!is.na(tmin)) %>%
select(ageb, date, tmp_hourly) %>%
# indicator for which hour is the min in a day
mutate(min_hour = map_dbl(tmp_hourly, ~ which.min(.x))) %>%
ggplot() +
geom_histogram(aes(x = min_hour), bins = 24)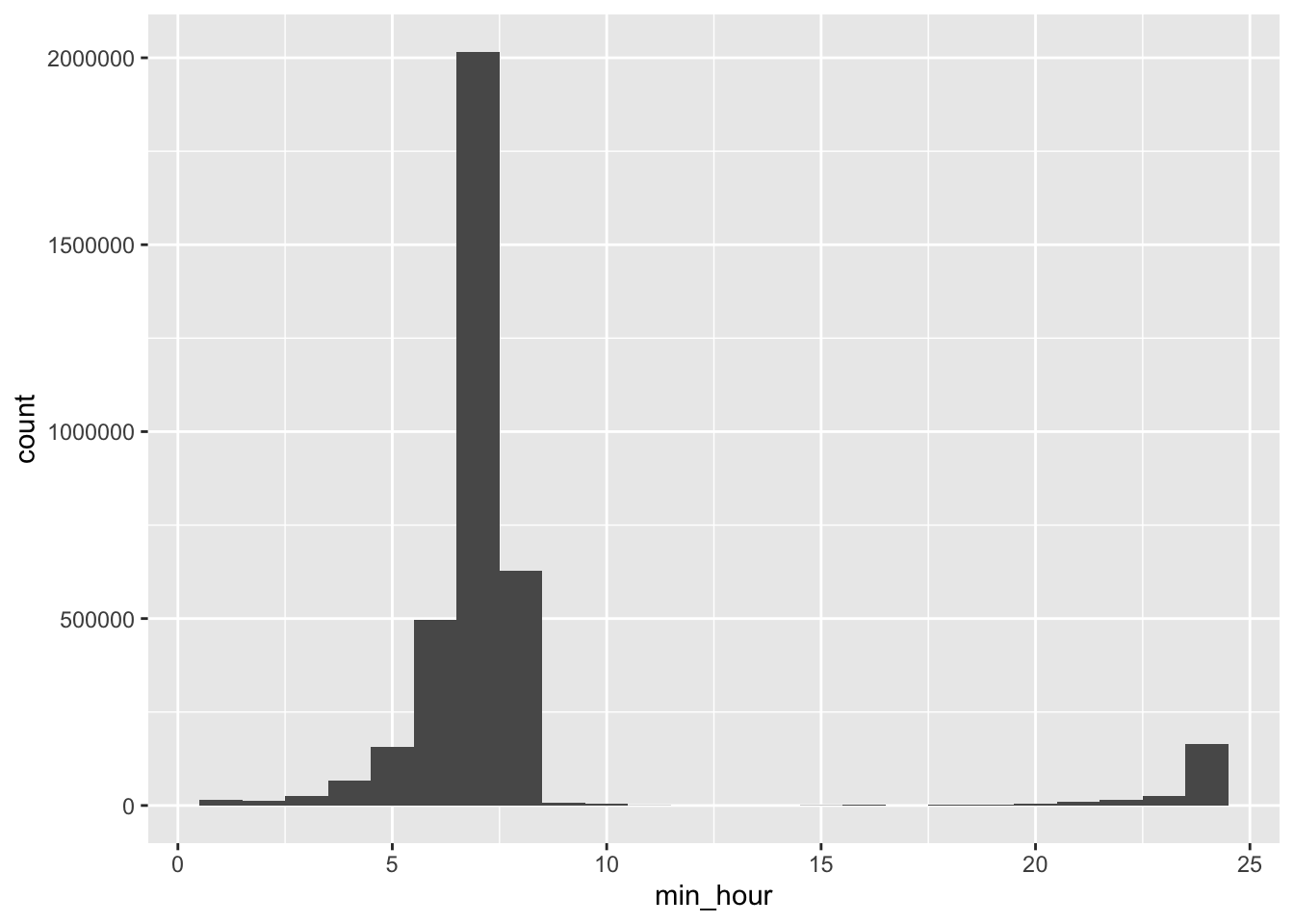
Since we have hourly measurements of temperature, we can calculate the average temperature during the day and during the night.
I define
- day temperature from 07:00 (hour 8) to 19:59 (hour 20)
- night temperature from 20:00 (hour 21) (the day before!) to 06:59 (hour 7)
So, there are 13 hours in a day, and 11 hours in the night
# I want to account for a different definition of night.
# now if i want the night to be LAGGED NIGHT TEMPERATURE:
# For a crime on Monday, night temperatures reflect Sunday 8:00 PM to Monday 6:59 AM.
data <- data %>%
mutate(tmean_day = map_dbl(tmp_hourly, ~ mean(.x[8:20])),
#this would be the "wrong i.e. not lagged" definition of night
#tmean_night = map_dbl(tmp_hourly, ~ mean(c(.x[21:24], .x[1:7]))),
tmean_lagged_night = map2_dbl(
lag(tmp_hourly, default = list(rep(NA, 24))), # Provide a default list of NAs
tmp_hourly,
~ if (!all(is.na(.x))) mean(c(.x[21:24], .y[1:7])) else NA_real_)
)Here’s the distribution of day and night temperatures.
data %>%
ggplot() +
geom_density(aes(x = tmean_day), color = "orange") +
geom_density(aes(x = tmean_lagged_night), color = "darkblue") +
theme_light()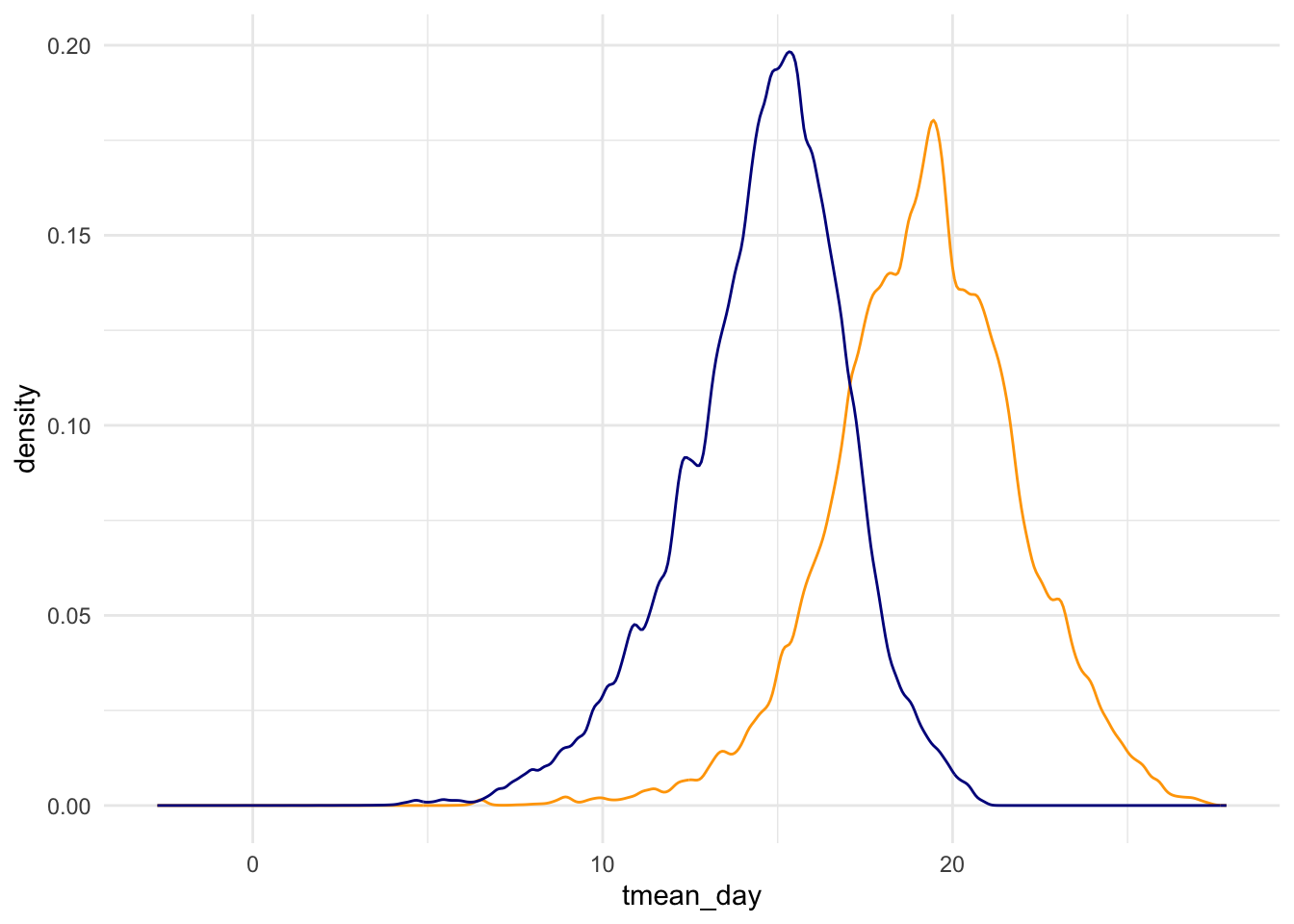
Now i can run the regs, separately for day and night temperatures, and jointly.
reg_day <- data %>%
fepois(reports_dv ~ tmean_day |
prec_quintile + rh_quintile + wsp_quintile +
ageb + year^month + day_of_week + day_of_year,
cluster = ~ ageb)
reg_night <- data %>%
fepois(reports_dv ~ tmean_lagged_night |
prec_quintile + rh_quintile + wsp_quintile +
ageb + year^month + day_of_week + day_of_year,
cluster = ~ ageb)
reg_day_night <- data %>%
fepois(reports_dv ~ tmean_day + tmean_lagged_night |
prec_quintile + rh_quintile + wsp_quintile +
ageb + year^month + day_of_week + day_of_year,
cluster = ~ ageb)Midnight temperature
Just to try, but doesnt add much
reg_midnight <- data %>%
mutate(midnight_temp = map_dbl(tmp_hourly, ~ .x[1])) %>%
fepois(reports_dv ~ midnight_temp |
prec_quintile + rh_quintile + wsp_quintile +
ageb + year^month + day_of_week + day_of_year,
cluster = ~ ageb)Export Table 1
These are the results we want to export:
etable(reg_linear_reports,
reg_day, reg_night, reg_day_night,
reg_max, reg_min, reg_min_max) %>%
kable()| reg_linear_reports | reg_day | reg_night | reg_day_night | reg_max | reg_min | reg_min_max | |
|---|---|---|---|---|---|---|---|
| Dependent Var.: | reports_dv | reports_dv | reports_dv | reports_dv | reports_dv | reports_dv | reports_dv |
| tmean | 0.0274*** (0.0034) | ||||||
| tmean_day | 0.0246*** (0.0030) | 0.0220*** (0.0041) | |||||
| tmean_lagged_night | 0.0188*** (0.0031) | 0.0039 (0.0042) | |||||
| tmax | 0.0198*** (0.0026) | 0.0159*** (0.0030) | |||||
| tmin | 0.0178*** (0.0029) | 0.0097** (0.0034) | |||||
| Fixed-Effects: | —————— | —————— | —————— | —————— | —————— | —————— | —————— |
| prec_quintile | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| rh_quintile | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| wsp_quintile | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| ageb | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| year-month | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| day_of_week | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| day_of_year | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| __________________ | __________________ | __________________ | __________________ | __________________ | __________________ | __________________ | __________________ |
| S.E.: Clustered | by: ageb | by: ageb | by: ageb | by: ageb | by: ageb | by: ageb | by: ageb |
| Observations | 3,624,826 | 3,624,815 | 3,624,809 | 3,624,797 | 3,624,826 | 3,624,826 | 3,624,826 |
| Squared Cor. | 0.01333 | 0.01333 | 0.01332 | 0.01333 | 0.01333 | 0.01332 | 0.01333 |
| Pseudo R2 | 0.05612 | 0.05612 | 0.05609 | 0.05612 | 0.05611 | 0.05609 | 0.05612 |
| BIC | 804,221.2 | 804,221.6 | 804,250.0 | 804,235.2 | 804,231.2 | 804,252.7 | 804,237.2 |
Export a latex with options. I have to reformat that table manually.
source(here("fixest_etable_options.R"))
etable(reg_linear_reports,
reg_day, reg_night, reg_day_night,
reg_max, reg_min, reg_min_max,
file = here("..", "output", "tables", "reg_diff_temperature_measures.tex"), replace = T,
adjustbox = TRUE, float = T,
title = "The Effect of Temperature on Reported DV",
label = "reg_diff_temperature_measures",
notes = "Standard errors clustered by neighborhood in parentheses. * denotes significance at the 10% level, ** at the 5% level, and *** at the 1% level. Panel A presents estimates from the baseline specification (Equation~\ref{eq_main_spec}), where temperature is measured as the mean of 24 hourly readings. Panel B replaces mean temperature with daytime (7:00–20:00) and nighttime (21:00–7:00) temperatures. Column (i) includes only daytime temperature; Column (ii) includes only nighttime temperature; Column (iii) includes both jointly. Panel C adopts the same approach using the minimum and maximum of the 24 hourly readings instead. The dependent variable is the count of reported domestic violence incidents per neighborhood per day."
)Plots for the quintiles of prec, hum, wsp
Now, we want to see the estimates for the quintiles. A table would be too cumbersome, so I do some plots:
reg_for_the_quintiles <- data %>%
fepois(reports_dv ~ tmean +
i(prec_quintile) + i(rh_quintile) + i(wsp_quintile) |
ageb + year^month + day_of_week + day_of_year,
cluster = ~ ageb)
prec_plot <-
iplot(reg_for_the_quintiles, only.params = T, i.select = 1)$prms %>%
ggplot(aes(x = estimate_names, y = estimate, ymin = ci_low, ymax = ci_high, group = 1)) +
geom_point(color = "black", size = 2) + geom_errorbar(color = "black", width = .2) +
geom_hline(yintercept = 0, alpha = .5) +
theme_light() +
labs(x = "Precipitation quintile", y = "Estimate and 95% CI")
hum_plot <-
iplot(reg_for_the_quintiles, only.params = T, i.select = 2)$prms %>%
ggplot(aes(x = estimate_names, y = estimate, ymin = ci_low, ymax = ci_high, group = 1)) +
geom_point(color = "black", size = 2) + geom_errorbar(color = "black", width = .2) +
geom_hline(yintercept = 0, alpha = .5) +
theme_light() +
labs(x = "Humidity quintile", y = NULL)
wsp_plot <-
iplot(reg_for_the_quintiles, only.params = T, i.select = 3)$prms %>%
ggplot(aes(x = estimate_names, y = estimate, ymin = ci_low, ymax = ci_high, group = 1)) +
geom_point(color = "black", size = 2) + geom_errorbar(color = "black", width = .2) +
geom_hline(yintercept = 0, alpha = .5) +
theme_light() +
labs(x = "Windspeed quintile", y = NULL)
prec_plot + hum_plot + wsp_plot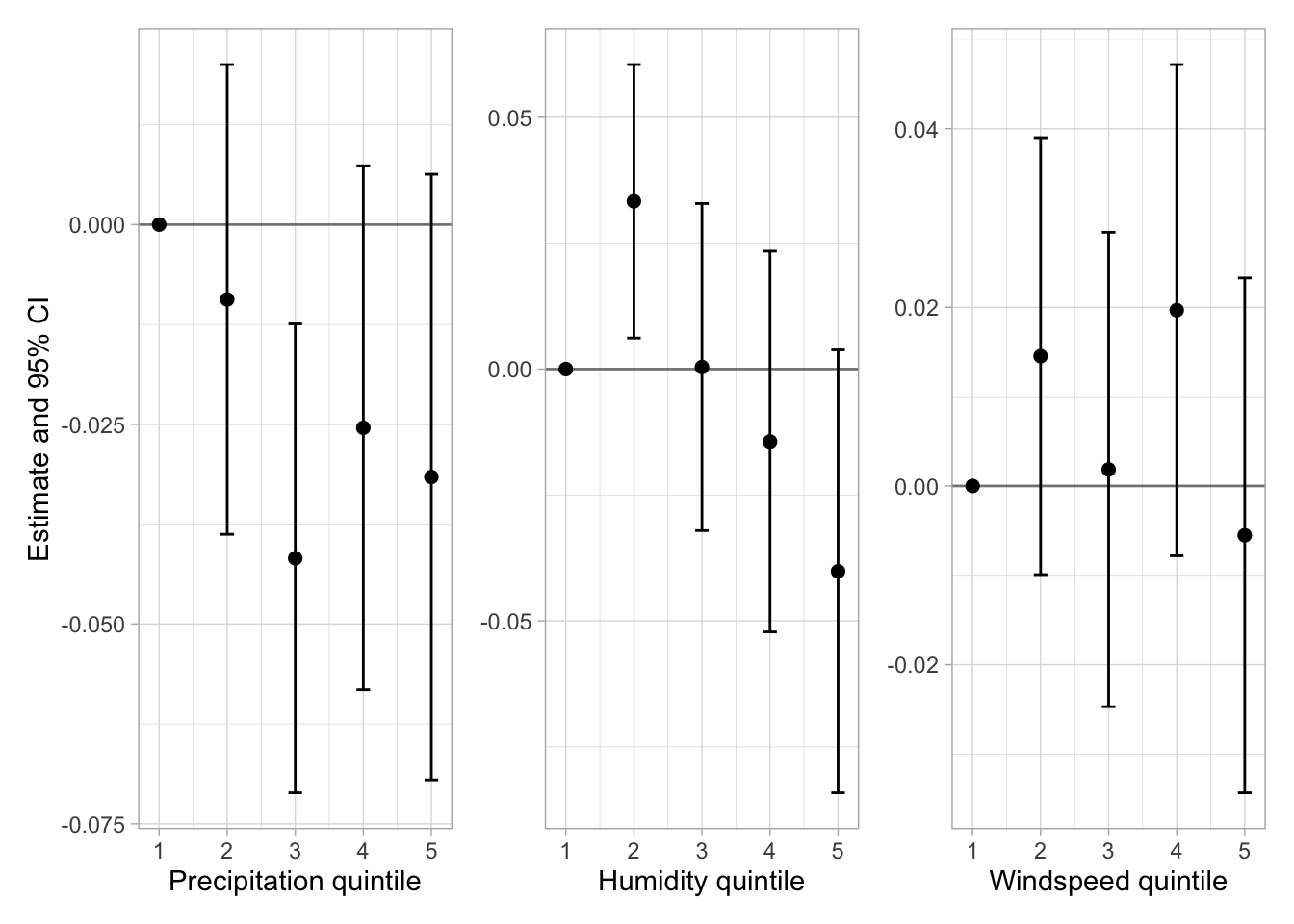
ggsave(path = here("..", "output", "figures"),
filename = "reg_quintiles.png",
width = 7, height = 2.3)Rendered on: 2025-07-27 01:21:38Total render time: 401.474 sec elapsed